Weekly Recap (Dec 2024, part 2)
Description
https://blog.stephenturner.us/p/weekly-recap-dec-2024-part-2
This week’s recap highlights a new way to turn Nextflow pipelines into web apps, DRAGEN for fast and accurate variant calling, machine-guided design of cell-type-targeting cis-regulatory elements, a Nextflow pipeline for identifying and classifying protein kinases, a new language model for single cell perturbations that integrates knowledge from literature, GeneCards, etc., and a new method for scalable protein design in a relaxed sequence space.
Others that caught my attention include commentary on improving bioinformatics software quality through teamwork, targeted nanopore sequencing for mitochondrial variant analysis, a review on plant conservation in the era of genome engineering, a de novo assembly tool for complex plant organelle genomes, learning to call copy number variants on low coverage ancient genomes, a near telomere-to-telomere phased reference assembly for the male mountain gorilla, a method for optimized germline and somatic variant detection across genome builds, a searchable large-scale web repository for bacterial genomes, and an integer programming framework for pangenome-based genome inference.
Audio generated with NotebookLM. (The hosts were very excited about this issue!)
Subscribe to Paired Ends (free) to get summaries like this delivered to your e-mail.
Deep dive
Cloudgene 3: Transforming Nextflow Pipelines into Powerful Web Services
Paper: Lukas Forer and Sebastian Schönherr. Cloudgene 3: Transforming Nextflow Pipelines into Powerful Web Services. bioRxiv, 2024. DOI: 10.1101/2024.10.27.620456.
I got to meet both Lukas and Sebastian in person at the Nextflow Summit. Lukas gave a talk on nf-test, while Sebastian gave a talk on the Michigan Imputation Server (MIS). MIS is implemented in Nextflow and driven using Cloudgene, and has helped over 12,000 researchers worldwide impute over 100 million samples. This paper describes Cloudgene for turning a Nextflow pipeline into a web service.
TL;DR: Cloudgene 3 provides a user-friendly platform to convert Nextflow pipelines into scalable web services, allowing scientists to deploy and run complex bioinformatics workflows without requiring web development expertise.
Summary: Cloudgene 3 addresses the challenge of deploying Nextflow pipelines as scalable web services, allowing researchers to leverage computational workflows without the need for technical setup or coding. The platform simplifies the transformation of Nextflow pipelines into “Cloudgene apps,” which include user-friendly interfaces and allow for seamless dataset management, job monitoring, and data security. By supporting features like workflow chaining and dataset integration, Cloudgene 3 enables collaborative and flexible use of pipelines across various scientific domains, from genomics to proteomics. This tool expands accessibility to complex analyses, facilitating data sharing and enhancing reproducibility, and has already been implemented in large-scale services like the Michigan Imputation Server. Its open accessibility and adaptable deployment model (cloud or local infrastructure) highlight its utility for bioinformatics workflows.
Methodological highlights:
* Converts Nextflow pipelines into web services with a few simple steps, creating portable “apps” that include metadata, input/output parameters, and multi-step workflows.
* Integrates real-time status updates and error handling for Nextflow tasks, leveraging a unique secret URL for each task to monitor progress.
* Supports cloud platforms and local installations, providing compatibility with engines like Slurm and AWS Batch and storage options like AWS S3.
New tools, data, and resources:
* Cloudgene 3 platform: Free platform available at cloudgene.io.
* Cloudgene 3 source code: https://github.com/genepi/cloudgene3.
Comprehensive genome analysis and variant detection at scale using DRAGEN
Paper: Behera, S., et al. Comprehensive genome analysis and variant detection at scale using DRAGEN. Nature Biotechnology, 2024. DOI: 10.1038/s41587-024-02382-1.
DRAGEN was a godsend in a previous job. I needed a turnkey variant calling solution that was fast. I bought an on-prem DRAGEN FPGA server, which was capable of taking you from FASTQ files to VCF in ~30 minutes for a 30X human whole genome. Illumina has previously published white papers on DRAGEN’s speed and accuracy. The publication in Nature Biotechnology engendered some interesting discussion online. On one hand, the paper was a pleasure to read, and the benchmarks are compelling and well done. On the other, the method isn’t available to explore, reproduce, understand in detail, or build upon. Which raises the question — should this have been a peer-reviewed publication in the scientific record? Or should this just have been another white paper? At some point “papers” hawking some new and improved closed source method are thinly veiled advertisements stamped with the approval of peer review. I think there should be some place in the scientific literature for papers like this describing a closed-source method, but where benchmarks are independently evaluated by a team of peer reviewers. I just don’t know what that looks like in the current landscape of peer reviewed papers versus a vendor’s white paper.
TL;DR: DRAGEN is a high-speed, highly accurate genomic analysis platform for variant detection, leveraging hardware acceleration, pangenome references, and machine learning. It outperforms traditional tools across variant types (SNVs, indels, SVs, CNVs, STRs) and is designed for large-scale, clinical genomics applications.
Summary: This study presents DRAGEN, a platform that uses accelerated hardware and sophisticated algorithms to enable comprehensive variant detection at unprecedented speed and accuracy. By integrating pangenome references and optimizing for all major variant classes, DRAGEN achieves high concordance in identifying complex and diverse genomic variants, even in challenging regions. Benchmarking across 3,202 genomes from the 1000 Genomes Project highlights DRAGEN’s scalability and its advantages over traditional methods like GATK and DeepVariant, especially for clinically relevant genes. The platform’s robust performance across SNVs, SVs, CNVs, and STRs allows for large-cohort analyses critical for population-scale genomics and clinical diagnostics, facilitating variant discovery in diseases with both common and rare genetic underpinnings.
Methodological highlights:
* Uses pangenome references to enhance alignment accuracy and variant detection across diverse populations.
* Optimized for rapid, parallel processing of SNVs, indels, CNVs, and STRs with an average processing time of ~30 minutes per genome.
* Employs machine learning-based filtering to reduce false positives and improve accuracy in variant calling.
* Integration of ExpansionHunter for STR analysis and specialized callers for pharmacogenomic variants (e.g., CYP2D6, SMN) ensures reliable detection in medically significant genes.
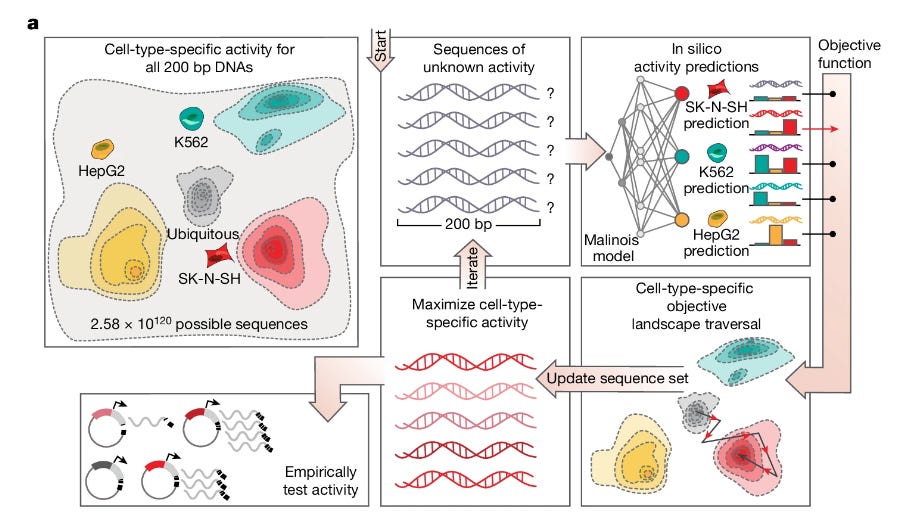
Machine-guided design of cell-type-targeting cis-regulatory elements
Paper: Gosai, S. J., et al. Machine-guided design of cell-type-targeting cis-regulatory elements. Nature, 2024. DOI: 10.1038/s41586-024-08070-z.
TL;DR: This paper introduces a platform for designing synthetic cis-regulatory elements (CREs) with programmed cell-type specificity using a deep-learning-based model called Malinois, combined with a computational design tool, CODA, and massively parallel reporter assays (MPRAs) for validation.
Summary: This study presents a framework for designing synthetic CREs that drive gene expression specifically in desired cell types. Using Malinois, a deep convolutional neural network trained on MPRA data from human cells, the researchers predict CRE activity and design synthetic elements targeting specific cell lines. The CODA (Computational Optimization of DNA Activity) platform then iteratively refines these designs to achieve high specificity, which is validated in vitro across multiple cell types and in vivo in mice and zebrafish. By outperforming natural CREs in specificity and robustness, these synthetic elements could significantly enhance targeted gene therapy approaches, especially by providing tools for precise gene expression control in therapeutic and research applications. The framework expands our capacity to engineer regulatory DNA for complex tissue-specific requirements, advancing possibilities for both biomedical research and gene therapy.
Methodological highlights:
* Malinois CNN model predicts cell-type-specific CRE activity directly from DNA sequences, validated with MPRA-based data in K5