Transcription Activators in Eukaryotes( CMB part 10)
Description
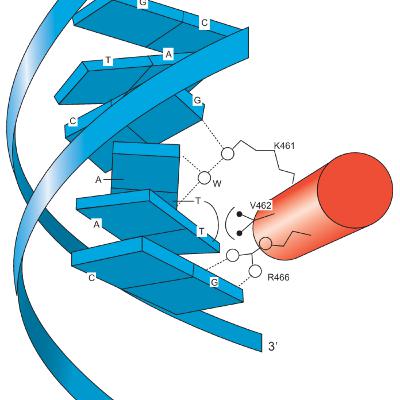
Eukaryotic activators consist of at least two domains: a DNA-binding domain and a transcription-activating domain. DNA-binding domains include motifs such as zinc modules, homeodomains, bZIP, or bHLH motifs. Transcription-activating domains can be acidic, glutamine-rich, or proline-rich. Zinc fingers are characterized by an antiparallel β-sheet followed by an α-helix. The β-sheet contains two cysteines, and the α-helix contains two histidines, which coordinate with a zinc ion to form the finger-shaped structure. This coordination facilitates specific recognition of the DNA target within the major groove.
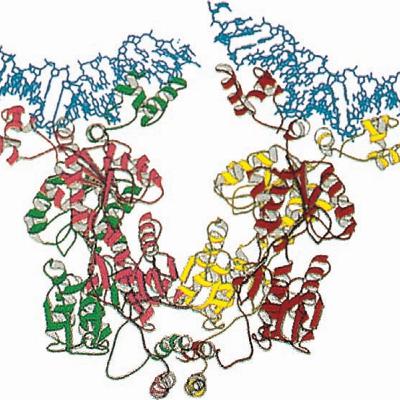
The DNA-binding motif of the GAL4 protein includes six cysteines that coordinate two zinc ions in a bimetal thiolate cluster. This motif features a short α-helix that extends into the DNA major groove, forming specific interactions. Additionally, the GAL4 monomer contains an α-helical dimerization motif that forms a parallel coiled coil with the α-helix of another GAL4 monomer.
Type I nuclear receptors are located in the cytoplasm, bound to other proteins. Upon binding their hormone ligands, these receptors release their cytoplasmic partners, translocate to the nucleus, bind to enhancers, and function as activators. A representative example is the glucocorticoid receptor, which contains a DNA-binding domain with two zinc modules. One module provides DNA-binding residues in a recognition α-helix, while the other facilitates protein-protein interactions for dimer formation. These zinc modules use four cysteine residues to complex the zinc ion, unlike classical zinc fingers, which use two cysteines and two histidines.
Homeodomains in eukaryotic activators contain a DNA-binding motif that operates similarly to the helix-turn-helix motifs in prokaryotes, where a recognition helix fits into the DNA major groove.