297-cSplotch: Inferring Cell-Type-Specific Spatial profile
Update: 2025-12-06
Description
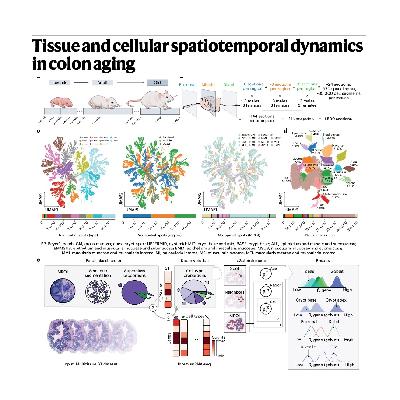
The source details the creation of a comprehensive spatiotemporal atlas of the mouse colon to characterize the complex tissue and molecular changes that occur during aging across various anatomical regions and life stages. This extensive resource was built by integrating data from roughly 1,500 tissue sections analyzed by spatial transcriptomics (ST) and over 400,000 profiles gathered through single nucleus RNA-sequencing (snRNA-seq). To synthesize this multimodal data, the authors developed a novel hierarchical Bayesian model called cSplotch, which accurately infers cell-type-specific gene expression by integrating molecular and histological information. The analysis identified significant molecular and cellular gradients along the longitudinal (proximal-distal) axis and the crypt axis that correspond to functional specialization within the organ. Temporal analysis revealed that aging is associated with a pervasive decline in secretory goblet cells and an expansion of progenitor cells, ultimately leading to a pervasive inflammatory state in the mucosal lining.
References:
- Daly A C, Cambuli F, Äijö T, et al. Tissue and cellular spatiotemporal dynamics in colon aging[J]. Nature biotechnology, 2025: 1-13.
Comments
In Channel