299-Limitations and Augmentation of Cell Embedding Metrics
Update: 2025-12-06
Description
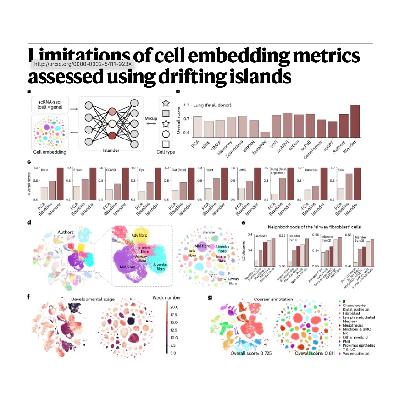
The source addresses the critical limitations in evaluating single-cell profile embeddings, which are widely used for characterizing cell types and states in biological investigations. To demonstrate the incompleteness of existing quality assessment metrics, the researchers introduce a model called Islander, a three-layer perceptron that outperforms established integration methods on standard metrics but severely distorts actual biological structures by creating separate cell "islands." This flaw reveals that current evaluation methods, such as those within the scIB framework, prioritize inter-batch mixing and tight cell clustering at the expense of preserving subtle biological continua or hierarchies. To provide a robust solution, the authors propose a new, complementary metric named scGraph, which evaluates how effectively an embedding preserves the similarities and relationships between cell types across different datasets. The text ultimately asserts that employing both the established scIB metrics and the novel scGraph is essential for a comprehensive evaluation framework that accurately measures quality while ensuring that meaningful biological variation is not obscured by overcorrection.
References:
- Wang H, Leskovec J, Regev A. Limitations of cell embedding metrics assessed using drifting islands[J]. Nature Biotechnology, 2025: 1-4.
Comments
In Channel