Discover Paper Talk
Paper Talk

297 Episodes
Reverse
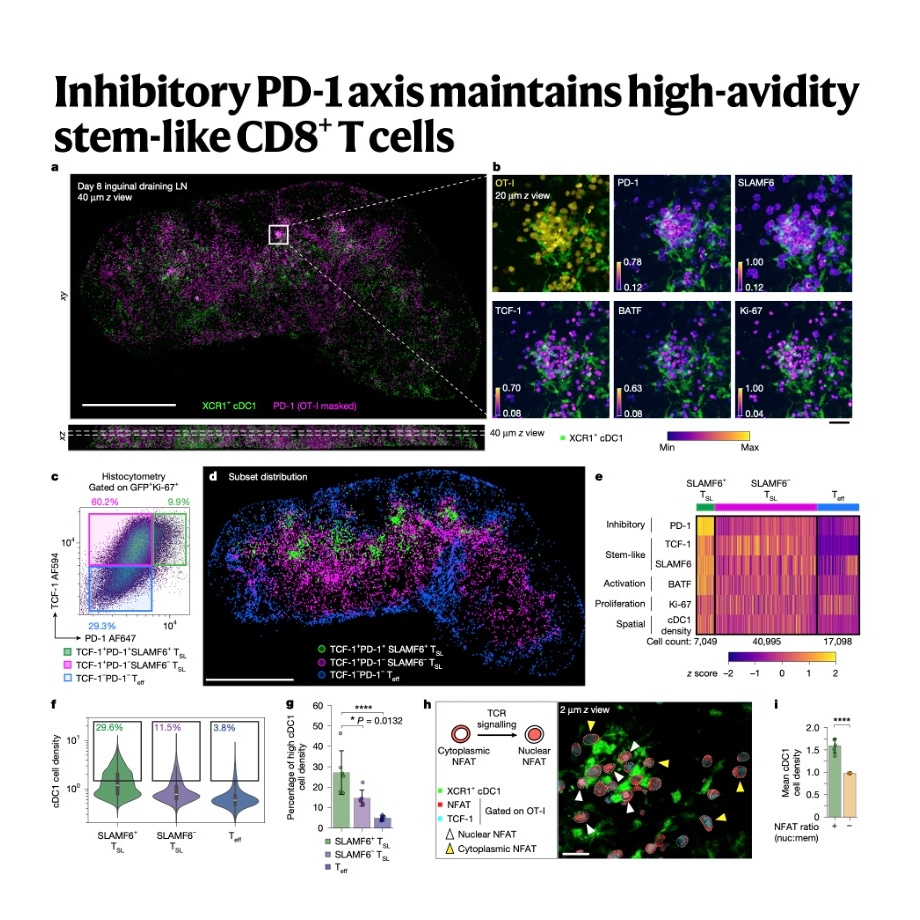
This research article investigates the mechanism by which stem-like CD8+ T cells (TSL), which are critical precursors for sustained anti-tumor responses, maintain their progenitor state despite chronic antigen exposure. The authors demonstrate that, contrary to traditional views, the inhibitory PD-1 pathway plays a crucial role by providing a negative-feedback loop that finely tunes the T-cell receptor (TCR) signaling intensity. This attenuation allows TSL cells with the highest TCR affinity to continuously proliferate and self-renew within specialized antigen-presentation niches in the tumor-draining lymph nodes. Crucially, checkpoint blockade that disables PD-1 inhibition disrupts this delicate balance, causing the high-affinity TSL population to undergo rapid terminal differentiation or cell death. The findings therefore suggest that while PD-1 blockade yields a powerful short-term anti-tumor effect, it may reduce long-term immunological maintenance by sacrificing the most potent progenitor cells.References: Hor J L, Schrom E C, Wong-Rolle A, et al. Inhibitory PD-1 axis maintains high-avidity stem-like CD8+ T cells[J]. Nature, 2025: 1-11.
The research utilizes an extensive dataset of animal harvests across 625 rural Amazonian localities to quantify the critical role of wild meat in sustaining the region's inhabitants. The study estimates that approximately 0.34 million metric tons of edible wild meat are extracted annually, supplying nearly half of the required daily intake of protein and iron, along with essential B vitamins, for over 10 million rural Amazonian peoples. This traditional food system, which relies heavily on mammals like peccaries and large rodents, is shown to be severely threatened by human-driven environmental pressures. Specifically, the availability of wild meat decreases drastically in areas experiencing extensive deforestation, higher human populations, and proximity to urban centers. The authors conclude that protecting the health and integrity of the Amazonian forest is paramount not only for biodiversity conservation but also for safeguarding the nutritional security and cultural autonomy of Indigenous and traditional communities.References: Antunes A P, de Araujo Lima Constantino P, Fa J E, et al. Healthy forests safeguard traditional wild meat food systems in Amazonia[J]. Nature, 2025: 1-9.
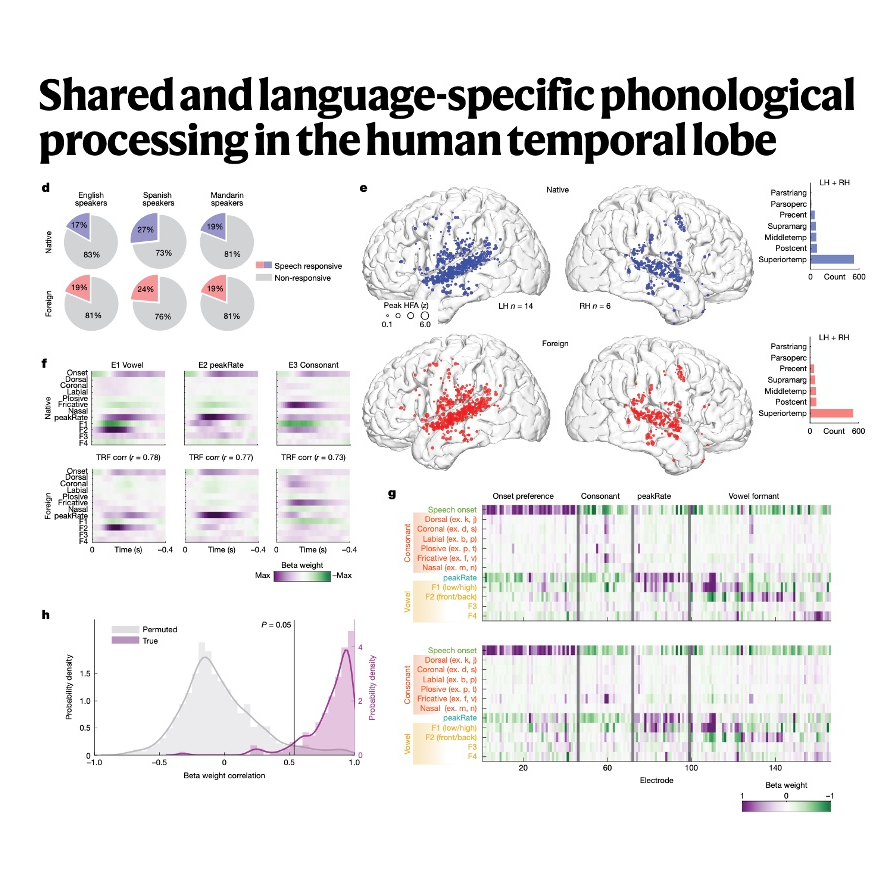
This research investigates the neural mechanisms in the human temporal lobe that differentiate between processing familiar and unfamiliar spoken languages, using high-density brain recordings (ECoG) in monolingual and bilingual speakers. The study determined that the superior temporal gyrus (STG) exhibits similar cortical activation and feature encoding for basic acoustic–phonetic features, such as vowels and consonants, regardless of whether the language is known or foreign. Critically, only when listening to their native language did the participants show enhanced neural encoding for higher-level linguistic structures, including word boundaries, word frequency, and specific sound sequence statistics. This strong dependency on experience indicates that the ability to segment continuous speech into individual words is a learned, language-specific skill localized to the STG. For bilingual individuals, this word-level processing capacity was available for both familiar languages, and the neural accuracy of extracting word-level information directly correlated with the speakers' proficiency in that language. These results propose a neurobiological model wherein the STG functions as an interface that integrates shared auditory processing with dynamic, experience-dependent word knowledge.References: Bhaya-Grossman I, Leonard M K, Zhang Y, et al. Shared and language-specific phonological processing in the human temporal lobe[J]. Nature, 2025: 1-12.
This paper details a comprehensive molecular analysis of intestinal fistulae, the penetrating lesions characteristic of Crohn’s disease, using advanced single-cell RNA sequencing and spatial transcriptomics (ST) on tissue samples from 92 individuals. The research defined a unique and abundant cell type called Fistula-Associated Fibroblasts (FAS cells), which are central to the persistence of the disease, unlike the related fibroblasts found in non-fistulating ulcers. These FAS cells organize into distinct spatial niches around the lesion, exhibiting powerful signatures associated with wound healing, tissue invasion, and extensive fibrotic remodeling of the extracellular matrix. The study further identified that specific transcription factors, including OSR2 and TWIST1, control these pathogenic fibroblast functions and promote dysregulated morphogenic signaling pathways. Overall, the findings reveal that the chronic inflammation and structural reprogramming seen in fistulae are maintained by complex, zone-specific macrophage–fibroblast cross-talk and a progression from superficial remodeling to deeper layer, stabilizing fibrosis.References: McGregor C, Qin X, Jagielowicz M, et al. Spatial fibroblast niches define Crohn’s fistulae[J]. Nature, 2025: 1-10.
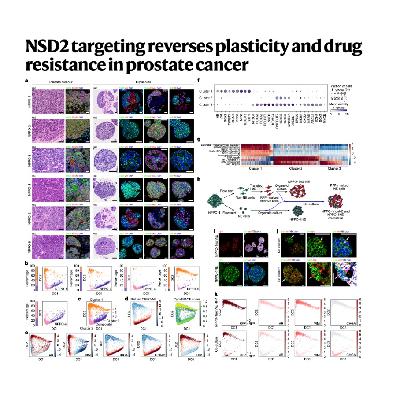
The paper details research into the molecular mechanisms driving resistance to androgen receptor (AR) pathway inhibitors, such as enzalutamide, in advanced Castration-Resistant Prostate Cancer (CRPC). The study specifically addresses the lineage plasticity through which tumors can transdifferentiate into the highly resistant neuroendocrine subtype (CRPC-NE). Investigators identified that the epigenetic regulator NSD2 is significantly upregulated in CRPC-NE, correlating with poor patient survival and driving this shift by increasing H3K36me2 histone marks. Using mouse and human organoid models, the research demonstrated that NSD2 targeting, either through genetic depletion or pharmacological inhibition, effectively reversed the neuroendocrine phenotype. This epigenetic reprogramming restored the tumors' AR expression and, consequently, their sensitivity to enzalutamide both in vitro and in vivo. Ultimately, the findings establish a strong preclinical basis for utilizing combined NSD2 and AR inhibition as a novel strategy to treat advanced and aggressive prostate cancer subtypes.References: Li J J, Vasciaveo A, Karagiannis D, et al. NSD2 targeting reverses plasticity and drug resistance in prostate cancer[J]. Nature, 2025: 1-11.
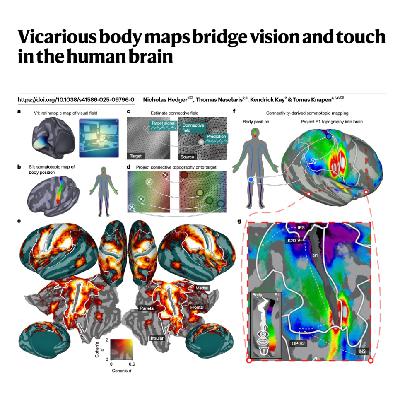
This paper introduces a computational model to investigate how the brain bridges visual input and the sense of touch, or somatosensation, focusing on the mechanism behind vicarious sensory experiences. Utilizing this model on fMRI data, the authors first confirmed the existence of intrinsic, organized somatotopic maps representing body-part tuning across the known somatosensory network during rest. A major finding was that this somatotopic organization expanded profoundly into the dorsolateral visual cortex during video viewing, demonstrating that visual input alone recruits spatially structured representations of touch. These newfound somatotopic maps in visual areas were shown to align with both visual field positions and categorical preferences for visual body parts. This revealed a pervasive cross-modal interface in brain organization that is ideally suited to translate visual impressions into body-referenced formats necessary for action and social understanding.References: Hedger N, Naselaris T, Kay K, et al. Vicarious body maps bridge vision and touch in the human brain[J]. Nature, 2025.
The research investigates a novel mechanism driving the progression of pancreatic ductal adenocarcinoma (PDAC) by demonstrating the presence of glutamatergic neuron-cancer pseudo-synapses between sensory nerve endings and cancer cells. This mechanism centers on the cancer cells' selective enrichment of the NMDA receptor subunit GRIN2D at the pseudo-synaptic sites, making them sensitive to neuron-derived glutamate. Activation of this glutamate-GRIN2D signaling pathway establishes a detrimental feedforward loop that significantly promotes tumor growth, invasiveness, and increased innervation in the pancreas. Furthermore, the study identifies that the loss of GRIN2D expression in cancer cells hinders tumorigenesis by suppressing the TGFA-EGFR signaling axis. Crucially, interfering with this glutamate-GRIN2D pathway dramatically enhanced survival in murine models, highlighting these peripheral cancer-neuron pseudo-synapses as a potential target for new oncological therapies.References: Ren L, Liu C, Çifcibaşı K, et al. Sensory neurons drive pancreatic cancer progression through glutamatergic neuron-cancer pseudo-synapses[J]. Cancer cell, 2025.
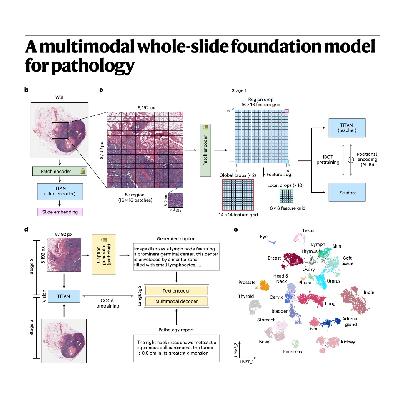
The paper describes the development and extensive evaluation of the TITAN (Transformer-based pathology Image and Text Alignment Network), a new multimodal foundation model for computational pathology. TITAN is pretrained on a massive dataset of over 335,000 whole-slide images (WSIs) using a combination of vision-only learning and subsequent vision-language alignment with both synthetic fine-grained captions and clinical pathology reports. The model utilizes a Vision Transformer architecture, enhanced with ALiBi positional encoding, to manage the immense scale and complexity inherent in gigapixel WSIs. Evaluations across diverse tasks, including cancer subtyping, molecular classification, and survival prediction, demonstrate that TITAN consistently outperforms prior slide foundation models, often by significant margins, even in data-limited settings such as rare cancer retrieval. Furthermore, its multimodal capabilities enable advanced functions like zero-shot visual-language classification, highly accurate cross-modal retrieval between slides and reports, and the automatic generation of clinical descriptions. Overall, the research demonstrates TITAN’s efficacy in producing powerful, general-purpose slide representations that are immediately applicable to complex clinical workflows without requiring task-specific fine-tuning.References: Ding T, Wagner S J, Song A H, et al. A multimodal whole-slide foundation model for pathology[J]. Nature Medicine, 2025: 1-13.
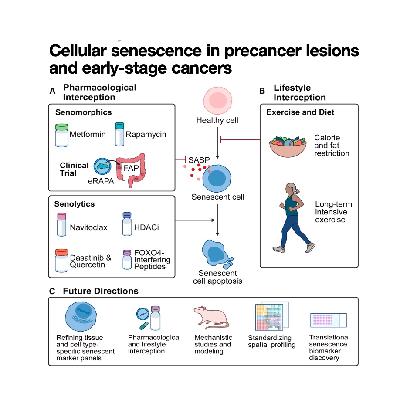
This commentary examines the ambivalent role of cellular senescence in the earliest stages of cancer, noting that it initially serves as a vital but transient tumor-suppressive barrier that eventually transitions into a pro-tumorigenic state. The authors analyze how this process extends beyond epithelial cells, highlighting the deleterious impact of senescence in stromal and immune cells that collectively reshape the precancer tissue microenvironment (PreTME). The text reviews promising precision interception strategies, including pharmacological agents (senolytics and senomorphics) and modifiable lifestyle interventions such as exercise and dietary restrictions. Furthermore, the source outlines critical hurdles for the field, primarily focusing on the necessity of developing reliable senescence markers and computational tools to effectively map and target these cells for prevention.References: Hoi X P, Stangis M M, Glass S E, et al. Cellular senescence in precancer lesions and early-stage cancers[J]. Cancer cell, 2025.
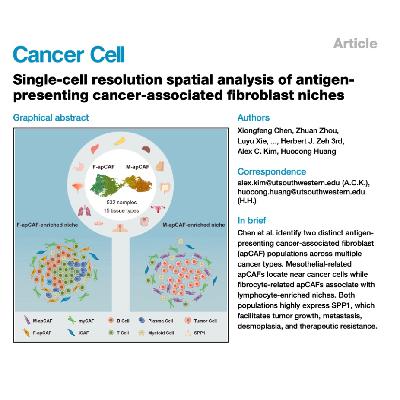
This article utilizes single-cell resolution spatial analysis across multiple solid tumors to characterize antigen-presenting cancer-associated fibroblasts (apCAFs). The study reveals two distinct apCAF populations—M-apCAFs (mesothelial-related) and F-apCAFs (fibrocyte-related)—that exhibit different spatial localizations within the tumor microenvironment. Specifically, the F-apCAFs are found predominantly in lymphocyte-enriched niches, while the M-apCAFs associate closely with cancer cells. A major functional finding is that both apCAF populations are the primary stromal source for secreted phosphoprotein 1 (SPP1), a protein known to drive tumor progression and therapy resistance. Further functional experiments in mouse models of peritoneal metastasis and pancreatic cancer demonstrate that targeting or knocking out host SPP1 significantly inhibits tumor growth and metastasis. The authors conclude that these spatial and molecular distinctions provide an important framework for understanding CAF heterogeneity and suggest that anti-SPP1 therapy may overcome therapeutic resistance.References: Chen X, Zhou Z, Xie L, et al. Single-cell resolution spatial analysis of antigen-presenting cancer-associated fibroblast niches[J]. Cancer Cell, 2025.
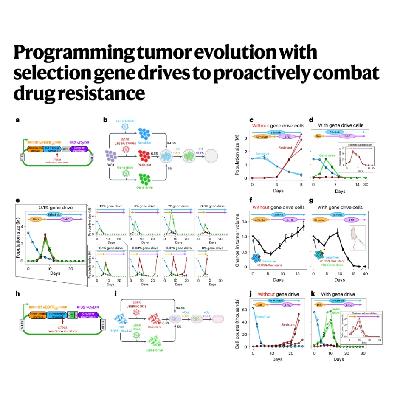
The article introduces a novel genetic engineering approach called selection gene drives to proactively combat the challenge of drug resistance in cancer treatment. This system is designed to redirect tumor evolution by modifying cancer cells with a dual-switch genetic circuit. The first switch provides a controllable fitness advantage under targeted therapy, promoting the engineered cells' expansion, while the second switch is a therapeutic payload that, when engaged, eliminates both the engineered and native drug-resistant cells via a bystander effect. Through the use of stochastic and spatial evolutionary models, the researchers established the necessary design criteria for this system, demonstrating in both in vitro and mouse models that this forward-engineering strategy can successfully eradicate diverse forms of pre-existing genetic resistance in tumors. The work highlights the modularity of the design and its potential to be a powerful framework for future evolution-guided anticancer therapy.References: Leighow S M, Reynolds J A, Sokirniy I, et al. Programming tumor evolution with selection gene drives to proactively combat drug resistance[J]. Nature Biotechnology, 2025, 43(5): 737-751.
The source addresses the critical limitations in evaluating single-cell profile embeddings, which are widely used for characterizing cell types and states in biological investigations. To demonstrate the incompleteness of existing quality assessment metrics, the researchers introduce a model called Islander, a three-layer perceptron that outperforms established integration methods on standard metrics but severely distorts actual biological structures by creating separate cell "islands." This flaw reveals that current evaluation methods, such as those within the scIB framework, prioritize inter-batch mixing and tight cell clustering at the expense of preserving subtle biological continua or hierarchies. To provide a robust solution, the authors propose a new, complementary metric named scGraph, which evaluates how effectively an embedding preserves the similarities and relationships between cell types across different datasets. The text ultimately asserts that employing both the established scIB metrics and the novel scGraph is essential for a comprehensive evaluation framework that accurately measures quality while ensuring that meaningful biological variation is not obscured by overcorrection.References: Wang H, Leskovec J, Regev A. Limitations of cell embedding metrics assessed using drifting islands[J]. Nature Biotechnology, 2025: 1-4.
The paper introduces a novel computational approach called the single-cell Polygenic Risk Score (scPRS), which is designed to overcome the limitations of conventional risk prediction by accounting for cellular and molecular heterogeneity in complex human diseases. This methodology integrates single-cell epigenome profiling, specifically scATAC-seq data, with a Graph Neural Network (GNN) to calculate genetic risk scores at the individual cell level. Researchers applied scPRS to four major conditions, including Type 2 Diabetes, Alzheimer Disease, and Hypertrophic Cardiomyopathy, consistently demonstrating superior predictive performance compared to established polygenic score methods. Critically, scPRS is capable of prioritizing and identifying disease-relevant cell types, such as specific pancreatic cells in T2D or microglia in AD. Furthermore, the model uncovers cell-type-specific genetic regulatory programs, allowing for the fine-mapping of causal risk variants and genes associated with disease pathogenesis. Experimental validation confirmed that genetic variations pinpointed by scPRS impact essential cellular functions, supporting the model's high resolution and biological interpretability.References: Zhang S, Shu H, Zhou J, et al. Single-cell polygenic risk scores dissect cellular and molecular heterogeneity of complex human diseases[J]. Nature Biotechnology, 2025: 1-17.
The source details the creation of a comprehensive spatiotemporal atlas of the mouse colon to characterize the complex tissue and molecular changes that occur during aging across various anatomical regions and life stages. This extensive resource was built by integrating data from roughly 1,500 tissue sections analyzed by spatial transcriptomics (ST) and over 400,000 profiles gathered through single nucleus RNA-sequencing (snRNA-seq). To synthesize this multimodal data, the authors developed a novel hierarchical Bayesian model called cSplotch, which accurately infers cell-type-specific gene expression by integrating molecular and histological information. The analysis identified significant molecular and cellular gradients along the longitudinal (proximal-distal) axis and the crypt axis that correspond to functional specialization within the organ. Temporal analysis revealed that aging is associated with a pervasive decline in secretory goblet cells and an expansion of progenitor cells, ultimately leading to a pervasive inflammatory state in the mucosal lining.References: Daly A C, Cambuli F, Äijö T, et al. Tissue and cellular spatiotemporal dynamics in colon aging[J]. Nature biotechnology, 2025: 1-13.
The paper introduces PRISM (profiling of RNA in situ through single-round imaging), a new high-multiplex fluorescent in situ hybridization (FISH) method designed to make spatial RNA imaging more accessible and efficient by overcoming the limitations of conventional microscopy. PRISM expands coding capacity by using color-intensity barcoding within a single round of staining, allowing for up to 64-plex detection without complex, expensive fluidics-dependent instrumentation. The article demonstrates PRISM’s robustness by applying it to create detailed spatial and quasi-3D atlases of diverse tissues, including mouse embryos, mouse brain sections, and human hepatocellular carcinoma (HCC). These applications reveal intricate biological details, such as cellular heterogeneity, developmental progression, and the complex microscale organization of the HCC tumor microenvironment.References: Chang T, Zhao S, Deng K, et al. High-plex spatial RNA imaging in one round with conventional microscopes using color-intensity barcodes[J]. Nature Biotechnology, 2025: 1-13.
The resource details the creation of a functional genomics atlas of over 148,000 regulatory regions in neural stem cells (NSCs) using ChIP-STARR-seq to understand gene regulation in neurodevelopment. Based on this atlas, the authors developed BRAIN-MAGNET, a convolutional neural network (CNN) that predicts non-coding regulatory element (NCRE) activity directly from DNA sequences. Critically, BRAIN-MAGNET is validated to prioritize functional non-coding variants linked to neurological diseases by identifying nucleotides essential for NCRE function, often outperforming existing variant prioritization tools in GWAS and rare disease contexts. The research employs rigorous experimental and computational methods to interpret the regulatory genome, including analyzing Transcription Factor (TF) motifs and epigenetic priming of enhancers.References: Deng R, Perenthaler E, Nikoncuk A, et al. BRAIN-MAGNET: A functional genomics atlas for interpretation of non-coding variants[J]. Cell, 2025.
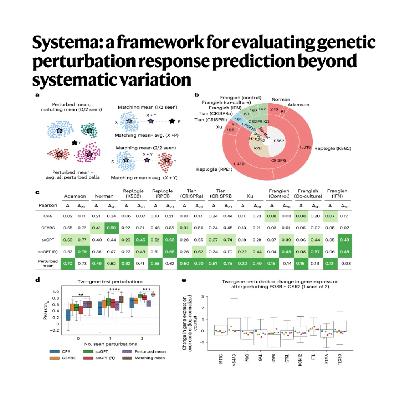
The article introduces Systema, a new evaluation framework designed to improve the prediction of transcriptional responses to genetic perturbations in functional genomics. The authors argue that existing computational methods for predicting these responses are often misleadingly successful because they predominantly capture systematic variation-consistent transcriptional differences between perturbed and control cells-rather than perturbation-specific effects. To address this overestimation of performance, Systema shifts the point of reference from the control cell centroid to the perturbed cell centroid, making evaluation more robust against these systemic biases. Using Systema, the research shows that predicting the effects of unseen genetic perturbations is more difficult than previously indicated by standard metrics, though some models, like fine-tuned scGPT, demonstrate a limited ability to infer coarse-grained, biologically meaningful perturbation effects. This work highlights the need for heterogeneous gene panels and reference-independent metrics to accurately assess the predictive power and biological utility of perturbation response models.References: Viñas Torné R, Wiatrak M, Piran Z, et al. Systema: a framework for evaluating genetic perturbation response prediction beyond systematic variation[J]. Nature Biotechnology, 2025: 1-10.
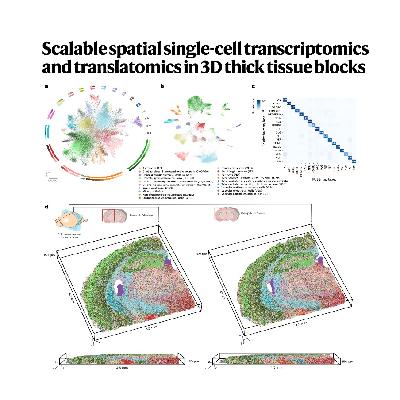
The paper introduces novel imaging platforms, Deep-STARmap and Deep-RIBOmap, designed for scalable, single-cell spatial transcriptomics and translatomics within thick, three-dimensional (3D) tissue blocks, overcoming limitations of traditional thin-sectioning methods. The researchers detail the technological advancements, including scalable probe synthesis and robust complementary DNA crosslinking, which enable the quantification of thousands of gene transcripts and their corresponding translation activities. The utility of these methods is demonstrated through comprehensive studies in mouse brain tissue, combining molecular profiling with neuronal morphology tracing, and in human cutaneous squamous cell carcinoma (cSCC) to analyze tumor–immune cell interactions in 3D. Ultimately, the new platforms provide a more accurate and quantitative understanding of gene expression and cellular organization within complex biological structures, offering significant potential for advancing neuroscience and oncology research.References: Sui X, Lo J A, Luo S, et al. Scalable spatial single-cell transcriptomics and translatomics in 3D thick tissue blocks[J]. Nature Methods, 2025: 1-11.
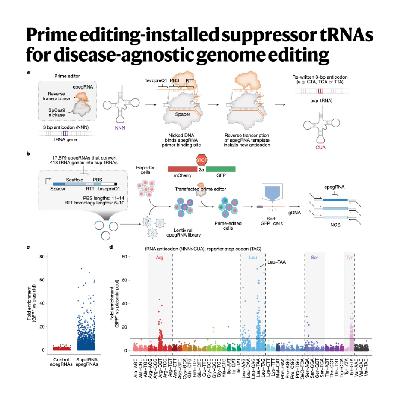
The paper introduces a disease-agnostic genome-editing strategy called Prime Editing-mediated Readthrough of Premature Termination Codons (PERT). This technique uses prime editing to permanently convert a redundant endogenous transfer RNA (tRNA) into an optimized suppressor tRNA (sup-tRNA) to rescue diseases caused by premature stop codons (nonsense mutations). The authors detail their iterative screening process to identify the most potent sup-tRNA variants and optimize the prime editing components for efficient, one-time genomic installation. Crucially, they demonstrate that this approach is effective in human cell models of diseases like Batten disease, Tay–Sachs disease, and cystic fibrosis, and it successfully rescues disease pathology in a mouse model of Hurler syndrome, all while showing no detectable toxicity or off-target effects on the transcriptome or proteome. PERT offers a generalized therapeutic solution that may bypass the need to develop a unique genome-editing treatment for every single pathogenic mutation.References: Pierce S E, Erwood S, Oye K, et al. Prime editing-installed suppressor tRNAs for disease-agnostic genome editing[J]. Nature, 2025: 1-12.
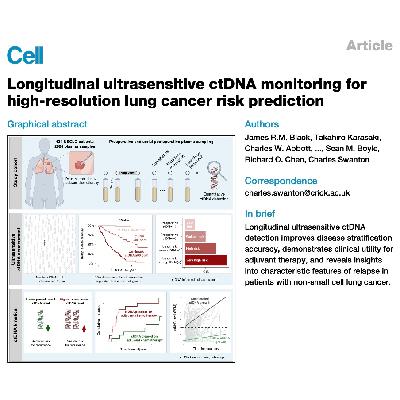
This article details a study focused on using longitudinal ultrasensitive circulating tumor DNA (ctDNA) monitoring to improve risk prediction and management for patients with non-small cell lung cancer (NSCLC). The research, based on the TRACERx study cohort, leverages a highly sensitive, tumor-informed ctDNA assay to detect molecular residual disease (MRD) both before and after surgery. Findings indicate that ctDNA detection levels, particularly those below 80 parts per million (ppm), are crucial for stratifying patient prognosis and identifying an intermediate-risk group. Furthermore, the kinetics of ctDNA clearance during adjuvant therapy are predictive of clinical outcome, suggesting a refined, ctDNA-guided approach for recommending and monitoring NSCLC treatments.References: Black J R M, Karasaki T, Abbott C W, et al. Longitudinal ultrasensitive ctDNA monitoring for high-resolution lung cancer risk prediction[J]. Cell, 2025.