292-PERT for Disease-Agnostic Nonsense Mutation Readthrough
Update: 2025-12-04
Description
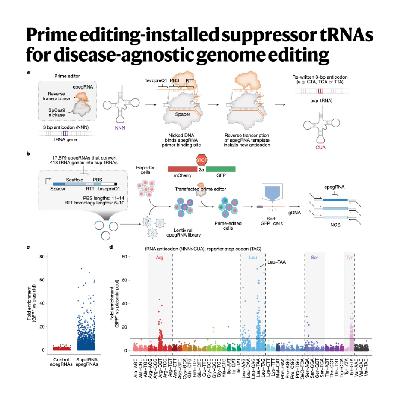
The paper introduces a disease-agnostic genome-editing strategy called Prime Editing-mediated Readthrough of Premature Termination Codons (PERT). This technique uses prime editing to permanently convert a redundant endogenous transfer RNA (tRNA) into an optimized suppressor tRNA (sup-tRNA) to rescue diseases caused by premature stop codons (nonsense mutations). The authors detail their iterative screening process to identify the most potent sup-tRNA variants and optimize the prime editing components for efficient, one-time genomic installation. Crucially, they demonstrate that this approach is effective in human cell models of diseases like Batten disease, Tay–Sachs disease, and cystic fibrosis, and it successfully rescues disease pathology in a mouse model of Hurler syndrome, all while showing no detectable toxicity or off-target effects on the transcriptome or proteome. PERT offers a generalized therapeutic solution that may bypass the need to develop a unique genome-editing treatment for every single pathogenic mutation.
References:
- Pierce S E, Erwood S, Oye K, et al. Prime editing-installed suppressor tRNAs for disease-agnostic genome editing[J]. Nature, 2025: 1-12.
Comments
In Channel