319-Spacedust: Discovery of Conserved Gene Clusters
Update: 2025-12-16
Description
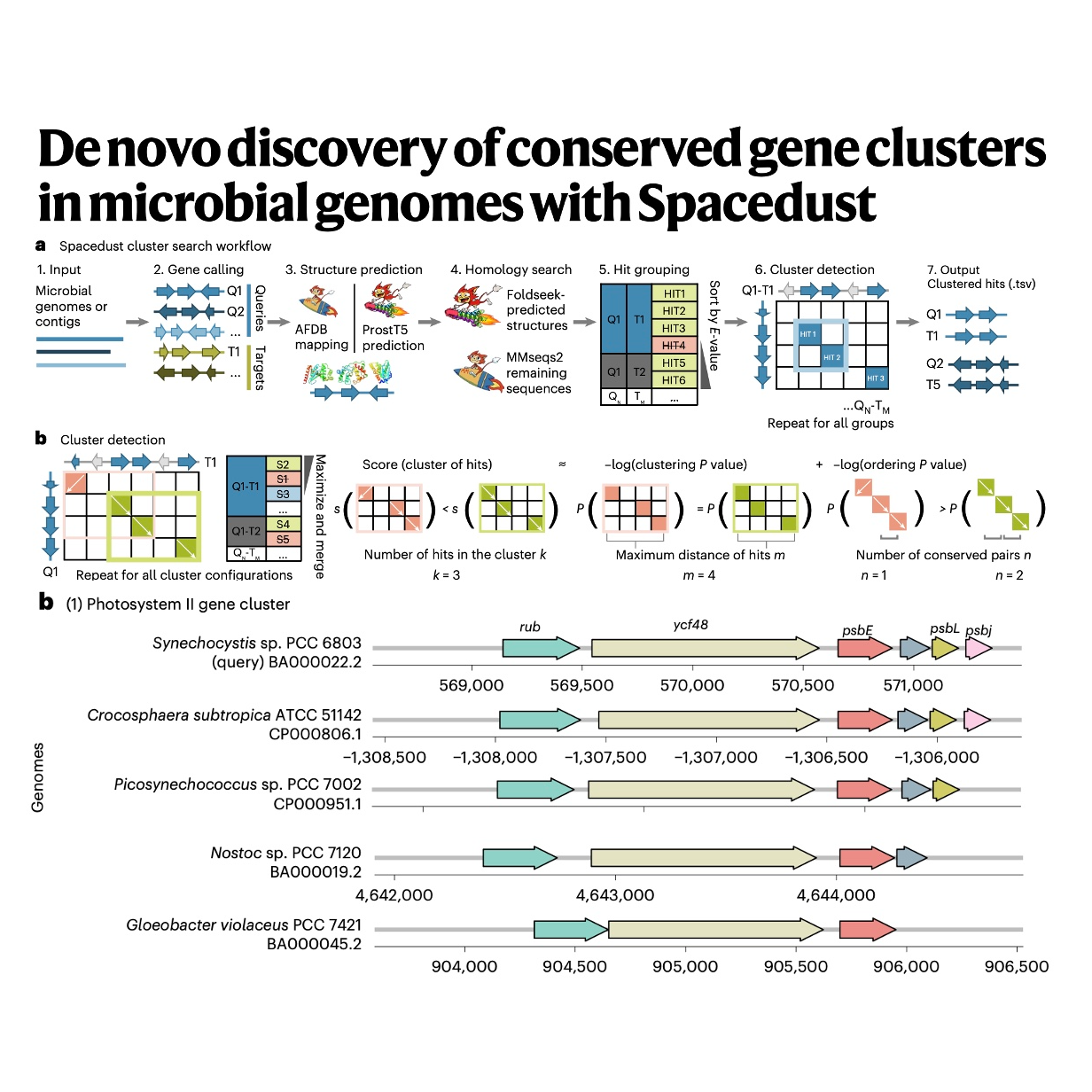
The paper introduces Spacedust, a novel computational tool created for the systematic, de novo discovery of conserved gene clusters (CGCs) in microbial and viral genomes, aiming to overcome the bottleneck of poor functional annotation in metagenomics. The software improves upon prior methods by employing highly sensitive protein structure comparison via Foldseek to find remote homologous matches that sequence-based methods often miss. To confirm the significance of these genetic neighborhoods, Spacedust utilizes novel statistical metrics based on clustering and ordering P values, allowing it to detect partial gene arrangement conservation. Validation demonstrated its high performance, as the tool identified CGCs containing 58% of all genes in a large bacterial dataset and successfully recovered 95% of antiviral defense systems annotated by dedicated software. Furthermore, Spacedust achieved higher accuracy (F1 scores) in detecting biosynthetic gene clusters (BGCs) compared to several specialized tools, establishing it as a rapid and powerful platform for large-scale genome annotation.
References:
- Zhang R, Mirdita M, Söding J. De novo discovery of conserved gene clusters in microbial genomes with Spacedust[J]. Nature Methods, 2025: 1-9.
Comments
In Channel