315-Unified Benchmark of TCR–Epitope Prediction Models
Update: 2025-12-14
Description
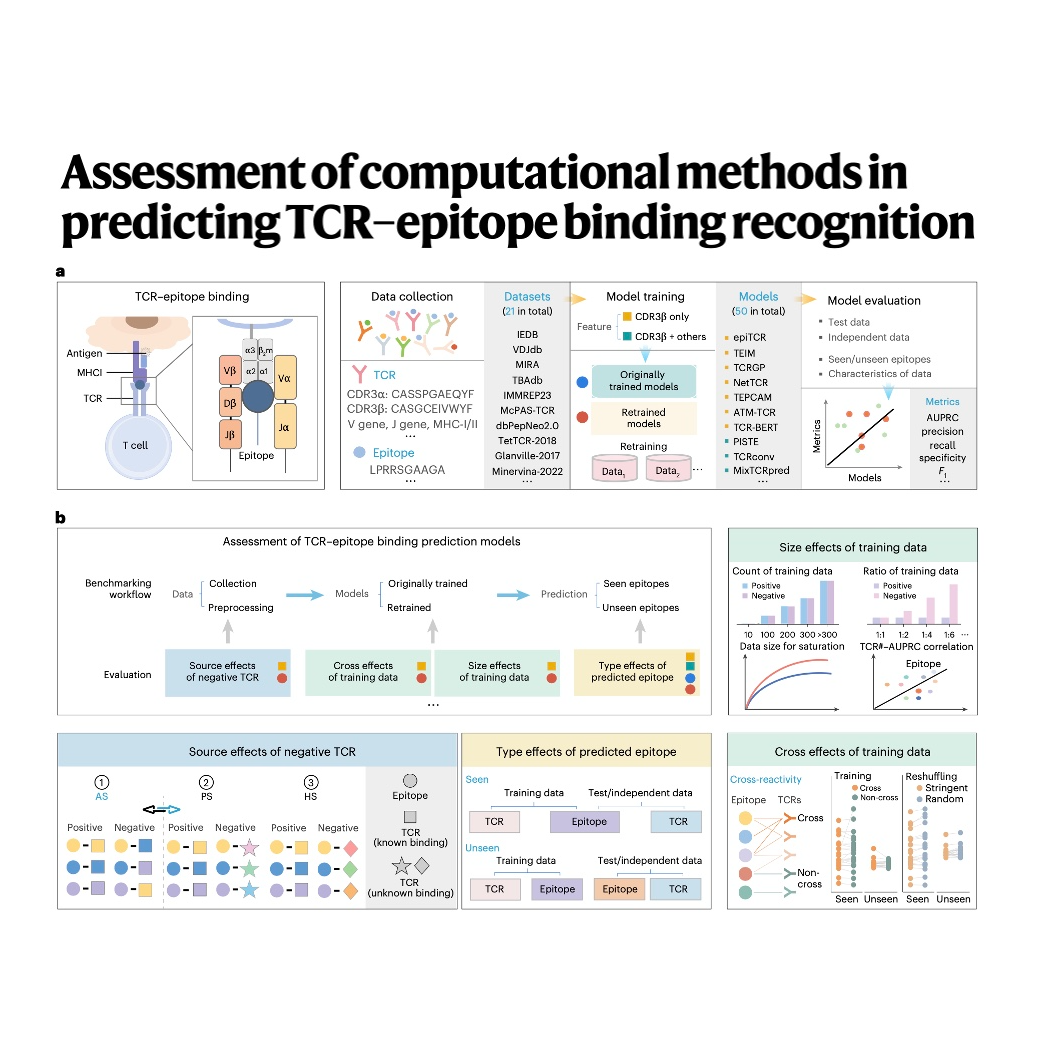
This study provides a thorough benchmark assessment of fifty cutting-edge computational methods designed for TCR-epitope binding recognition. The authors implemented a standardized methodology, including comprehensive retraining across unified datasets, to evaluate model effectiveness and identify key influencing factors. The analysis reveals a significant challenge in the field: most models struggle significantly to generalize when predicting interactions involving unseen epitopes, often resulting in near-random performance. However, accuracy generally improved when models utilized larger training datasets and incorporated multiple features beyond just the CDR3$eta$ sequence, such as MHC class and $\alphaeta$ chain information. Crucially, the research determined that model reliability is highly dependent on the source of negative control TCRs and the stringent requirement for independent test sets to provide unbiased results.
References:
- Lu Y, Wang Y, Xu M, et al. Assessment of computational methods in predicting TCR–epitope binding recognition[J]. Nature Methods, 2025: 1-12.
Comments
In Channel